That headline catches your eye, doesn’t it?
We’ve seen such claims made in popular media such as the March 2010 Fury as EU approves GM potato: Critics claim plant could spread antibiotic-resistant diseases to humans in the Independent: “Opponents fear bacteria inside the guts of animals fed the GM potato – which can cause human diseases – may develop resistance to antibiotics.” Groups that actively work against deregulation of genetically engineered crops have been making such claims for years.
We’ve also seen these claims in peer-reviewed journals (although, far less frequently than in non-peer reviewed media and reports). For example, in the February 2009 issue of Critical Reviews in Food Science and Nutrition, the review Health Risks of Genetically Modified Foods: “An area of concern focuses on the possibility that antibiotic resistance genes used as markers in transgenic crops may be horizontally transferred to pathogenic gut bacteria, thereby reducing the effectiveness of antimicrobial therapy.”
Are antibiotic marker genes in genetically engineered crops really a risk to human health? Many people have raised this question and there seems to be a lot of confusion about the issue. It’s time to look into the risks and reasons more deeply.
What are antibiotic resistance genes for?
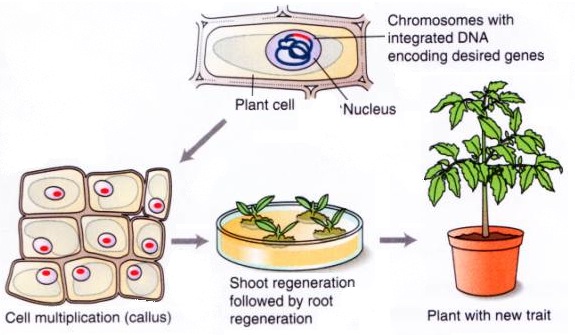
In order to understand why these genes are used, we have to look a little at the process of genetic engineering. For some plant types, including corn and rice, immature seeds are dissected to expose the developing embryo. Pieces of carrot roots can be transformed, as can the leaves of tobacco.
The desired genes are transferred into the plant cells with either a gene gun or Agrobacterium. The plant cells are then grown up into whole plants in petri dishes, with the help of plant hormones. The process is similar to other asexual plant propagation techniques, but much smaller!
Not every cell receives the gene of interest, however, so researchers need a way to find the cells that have it. Enter antibiotic resistance genes. If the cells are transformed with the gene of interest and an antibiotic resistance gene, the appropriate antibiotic can be added to the media in the petri dish so that any cells that didn’t get the genes will die. The antibiotic resistance gene is being used as a selectable marker, since it allows the researcher to select only the desired cells.
Of course, just because these genes are useful doesn’t mean that they are safe or that they should be used. What does the research tell us?
Risk: Gene transfer from plants to bacteria
Fear of antibiotic resistance markers is mainly due to fear of gene transfer from genetically modified plants to bacteria in the soil or bacteria in human or animal guts.
There are at least two reasons why this fear is unwarranted. First, soil and gut bacteria naturally contain a variety of antibiotic resistance genes without any human intervention. Second, transfer of genes from a plant to a bacterium is extremely unlikely.
Natural antibiotic resistance genes
Life for a bacterium isn’t easy. They have to compete fiercely for resources, so it’s not surprising that some bacteria have evolved to produce poison that kills their competitors: antibiotics. The producers of these antibiotics also evolved antibiotic resistance mechanisms so they could survive their own weapons. Additionally, bacteria develop resistance to antibacterial compounds in the environment.
Often, antibiotic resistance is conferred by a single gene. Any bacteria that can find that resistance gene and use it have an advantage. Consequently, antibiotic resistance genes are widespread in natural environments. When humans intervene, using antibiotics in ways that encourage development of resistance in bacteria, that resistance is passed around even faster (no GMOs needed). For some information on how humans, check out the CDC’s pages on antibiotic resistance.
Gene swapping
Genetically modified crops: methodology, benefits, regulation and public concerns, a 2000 review in the British Medical Bulletin has a summary of the risks of horizontal gene transfer from genetically modified crops:
…horizontal transfer of a gene from ingested plant material to bacteria has never been demonstrated, and there is no indication that it has ever occurred during evolution. The probability that it could occur is, therefore, considered to be so low that it is not relevant when compared with the natural occurrence of antibiotic resistance genes.
They sound awfully confident, don’t they?
Bacteria (prokaryotes) are fairly promiscuous when it comes to genes. Many types of bacteria (not all) have the ability to take up DNA from the environment and from other bacteria and integrate it into their own genome during parts or all of their life cycle. This process is called horizontal gene transfer, and the bacteria that have this ability are called competent. Since they have this ability, it makes sense to worry about bacteria picking up antibiotic resistance genes (and other genes as well) from other organisms, including genetically modified crops.
Interestingly, eukaryotes (multicellular organisms like plants and people) also have the ability to take up DNA into their genomes. For example, the August 2007 Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes shows transfer of the entire genome from endosymbiotic bacteria into their hosts’ genomes. A more recent example appeared in January 2010 in the New York Times: Hunting Fossil Viruses in Human DNA. An entire virus genome resides in the human genome, and has been passed down from our simian ancestors.
So we know that bacteria can swap DNA and that eukaryotes can take up DNA from bacteria and viruses. Can prokaryotes take up DNA from eukaryotes?
It doesn’t look like it.
The European Food Safety Authority has the following to say about the subject in their excellent 2009 Statement of EFSA on the consolidated presentation of opinions on the use of antibiotic resistance genes as marker genes in genetically modified plants (section 2.1.1.2., edited slightly for clarity):
While many studies support the evolutionary significance of horizontal gene transfer between bacteria, eukaryotic genes in prokaryotic genomes are a rarity. There is no definitive report of DNA transfer from eukaryotes to bacteria.
As of 24 September 2008 the public genome databases included more than 750 completed prokaryotic genomes. In the first annotation of the putative genes there are frequent cases where closest matches are found with eukaryotic genes, but these preliminary results have not manifested into demonstrations of horizontal gene transfer from eukaryotes to prokaryotes, as judged by the scientific publications interpreting the genomic sequencing data. For one functional gene (phosphoglucose isomerase), phylogenetic analyses indicated that the gene might have been transferred from a eukaryote to bacteria. The transfer was estimated to have happened approximately 500 million years ago.
Multiple studies have found that bacteria can take up eukaryote DNA, but only in certain conditions, where the researchers used a sort of genetic trick called homologous recombination.
In short, homologous recombination can occur when the ends of the donor DNA have sequences similar to part of the acceptor DNA. The homologous sequences can bind together, and in the next round of replication, the donor DNA can be integrated. In studies that aimed to find evidence of transfer of DNA from eukaryotes to prokaryotes without genetic tricks, none was found.
Table 1 of the EFSA Statement lists all studies prior to its publishing that examined horizontal gene transfer in bacteria (25 studies in all). Of the 18 studies that looked for gene transfer with homologous recombination, 15 found gene transfer and 3 did not. Of the 16 studies that looked for gene transfer without homologous recombination, no evidence of gene transfer was found.
In short, there are many DNA sequences that look like eukaryote genes in prokaryote genomes, but so far only one has been found that might be an actual functional gene. All evidence to date shows that gene transfer from eukaryotes to prokaryotes can only occur when homologous DNA sequences are present in donor and acceptor genomes. The lack of evidence for horizontal gene transfer in the wild suggests that there are some sort of barriers to gene transfer from eukaryotes to prokaryotes.
Barriers to gene swapping
Prokaryotic DNA and eukaryotic DNA are sort of like different computer languages. In fact, each species has slightly different ways of “personalizing” its own DNA with things like methylation and other DNA modifications, different codon preference, post translational modification of RNA, and whether or not introns are present. Eukaryotic DNA is so different from prokaryotic DNA that the bacteria just can’t take it up and use it as they would other bacterial DNA.
Additionally, even if all of the barriers to gene uptake occur and all the barriers to gene expression are overcome, the likelihood that the gene will confer a positive trait for the bacterium is low. Most eukaryotic genes aren’t going to be helpful for a prokaryote, such that the few useful genes are few and far between.
Even if a bacterium was able to uptake and express an antibiotic resistance gene from a genetically engineered plant, there would have to be selective pressure (i.e. an environment that included the antibiotic that the gene conferred resistance to) in order for the gene to be maintained in a bacterial colony. For more information about barriers to gene swapping, check out the EFSA Statement.
Avoiding the unlikely
Despite the fact that horizontal gene transfer from eukaryotes to prokaryotes is so unlikely (the only known example was estimated to have happened 500 million years ago), there are still precautions that can be taken to make it even more unlikely. For example, if antibiotic resistance genes are used as selectable markers in genetically modified organisms, researchers can avoid using sequences with homology to known bacterial genomes, they can be sure to only use antibiotic resistance genes that include features that make the gene unusable in bacteria, and they can avoid using promoters that are active in bacteria, just to name a few.
Alternative markers
Another option is to find alternative marker genes and alternative strategies. Herbicide resistance genes are an alternative selectable marker. Visible markers like GFP or GUS are screenable markers. Marker genes can be bred out, meaning that the final plant line will not contain the marker gene. Finally, it is possible to use no marker genes at all, but that does require far more screening of adult plants which can add expense and time to any project.
Peer-reviewed works cited
- Dona A, & Arvanitoyannis I (2009). Health Risks of Genetically Modified Foods Critical Reviews in Food Science and Nutrition, 49 (2), 164-175 DOI: 10.1080/10408390701855993
- Halford NG & Shewry PR (2000). Genetically modified crops: methodology, benefits, regulation and public concerns. British medical bulletin, 56 (1), 62-73 PMID: 10885105
- Hotopp JC, et al. (2007). Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science (New York, N.Y.), 317 (5845), 1753-6 PMID: 17761848
Other works cited
- Collins JD, et al. (2009). Statement of EFSA on the consolidated presentation of opinions on the use of antibiotic resistance genes as marker genes in genetically modified plants. European Food Safety Authority.
- Hickman M & Roberts G (2010). Fury as EU approves GM potato: Critics claim plant could spread antibiotic-resistant diseases to humans. The Independent.
- Zimmer, C (2010). Hunting Fossil Viruses in Human DNA. New York Times.
Note: This work was originally posted at Scientific Blogging as an entry in their 2010 Scientific Blogging Contest, and was an Editor’s Selection.