Written by Alma Laney
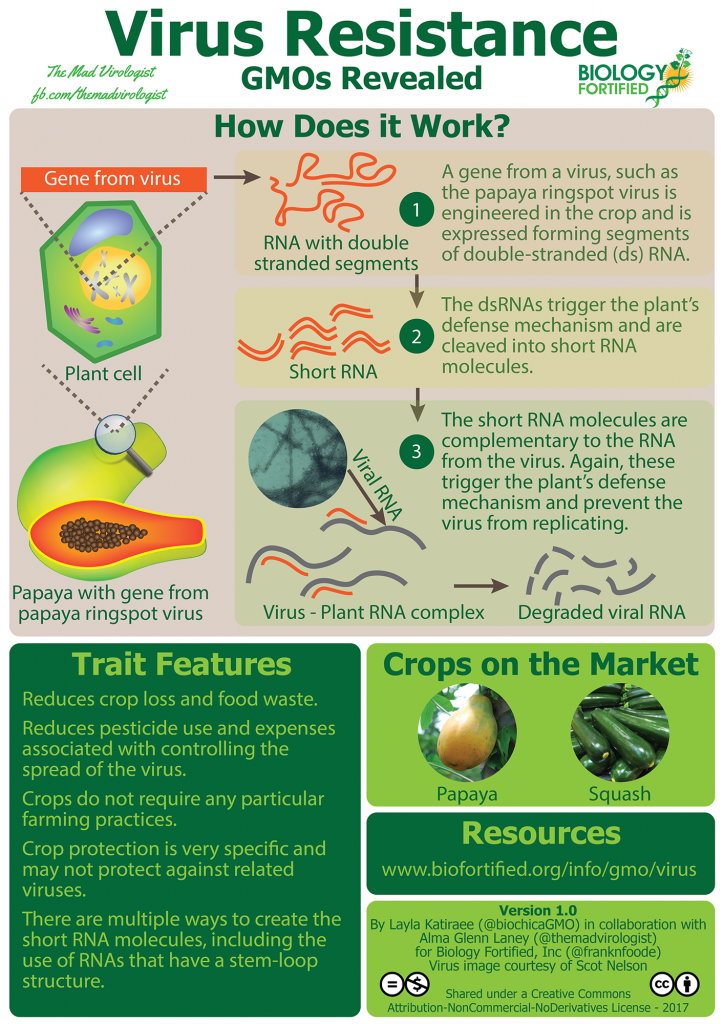
I’m Alma Laney from The Mad Virologist, and I work with plant viruses and the insects that help them spread. I’m happy to announce that Biology Fortified just published a new infographic that I helped develop with Layla Katiraee and the rest of the team here at Biology Fortified to help explain how virus resistance is created in genetically engineered crops.
The GMOs Revealed infographic on Virus Resistance distills a lot of information about how to engineer this trait into plants, with papayas and squash as the current examples on the market. There is really a lot more to this issue than these two examples, so I wanted to give a detailed overview explaining the challenges that farmers face with viral plant diseases, methods of control, and approaches to engineering resistance. Go to the infographic page for the short version, but if that’s not enough to ‘inoculate’ your curious mind, read on!
Introduction
Plant viruses can be serious pathogens in crops as they can cause anywhere from minor losses to a total loss. Viruses can infect crops in a number of ways ranging from being transmitted by contaminated tools, seed and pollen infections, infection of tubers or other vegetatively propagated material, and by arthropod vectors1 (what mites and insects are collectively known as). Most plant viruses are transmitted to crops via arthropod vectors. Plant viruses are of concern to farmers because once they get into a crop, all you can do is try and prevent their spread. Because of this, control strategies focus on preventing the introduction and spread of these viruses.
Control of plant viruses
There are several strategies that farmers use to prevent and control viral infections:
- Use certified seed or plants, which have been tested for known pathogens. Lots containing pathogens are rejected by the company.
- Control weeds around the fields as weeds can harbor both viruses and their vectors, serving as a source of inoculum for the field.
- Limit the spread of soil and the use of dirty implements. Some plant viruses can be carried by infested soil and contaminated implements can transfer the virus to healthy plants. There are many disinfectants that can be used including dilute bleach and milk solutions.
- Use seed treatments and/or spraying insecticides on the crop. Since most plant viruses are transmitted by arthropod vectors, this can be an effective strategy but it does not work in cases where the viruses are transmitted quickly (in non-persistent or semi-persistent transmission; see Table 1).
- A control strategy that isn’t always effective is to use a mild strain of a virus that is inoculated onto a plant. The mild strain can induce resistance to more severe strains, but this is problematic as mixed infections with other viruses can cause severe disease, the mild strain could become more severe, the mild strain would still cause economic losses, or the mild strain just may not work. This strategy is called cross protection.
- Once a virus is in a field, farmers can take action by roguing symptomatic plants (removal of infected plants followed by destroying them). This can be a costly measure and is used in cases where the type of virus can be transmitted quickly.
Table 1: The types of virus transmission2 detailing how long it takes to acquire a virus, how long it takes before the virus can be transmitted, and an example of each.
| Transmission type | Virus acquisition | Virus transmission | Example |
| Non-persistent | As little as a few seconds | A few minutes | Papaya ringspot potyvirus |
| Semi-persistent | Several minutes | Minutes to hours | Cauliflower mosaic caulimovirus |
| Circulative, non-propagative | Minutes to hours | Hours to days | Barley yellow dwarf luteovirus-PAV |
| Circulative, propagative | Minutes to hours | Up to a few weeks | Tomato spotted wilt tospovirus |
However, one of the best strategies is to engineer resistance to plant viruses. There are several examples of plant virus resistance genes that can be found in the germplasm (a collection of seed and/or plant tissue reflecting a variety of genotypes). However, for some crops, there are no known resistance genes or if they exist, they are found in wild relatives and attempts to introgress (move a trait into a crop plant by conventional breeding) the genes have not been successful. This is where genetic engineering can provide a solution. Genetically engineered plant virus resistance induces two different forms of resistance and one of the methods can use two different approaches.
Transgenic approach to virus control
Early on, virologists transformed plants with the complete virus coat protein gene, which forms the shell of the virus to protect the genetic material. It was found in the case of Tobacco mosaic tobamovirus (TMV), that over-expression of the coat protein gene led to virus resistance because the excess coat protein interfered with the ability of the virus to complete its lifecycle and move systemically in the plant3. Based on this, transgenics for other viruses were generated including Papaya ringspot potyvirus (PRSV), Cucumber mosaic cucumovirus (CMV), Zucchini yellow mosaic potyvirus (ZYMV), and Watermelon mosaic potyvirus (WMV). However, later research uncovered a second way that transgenics can induce resistance to plant viruses: triggering the RNA silencing (RNAi) pathway found in plants.
The discovery of RNAi was revolutionary. It has led to other control strategies as well as providing a powerful tool for functional analysis of genes. In plants, the RNAi pathway serves as a type of immune system for plants to target pathogens4, including viruses. There are multiple ways that the RNAi pathway can be triggered:
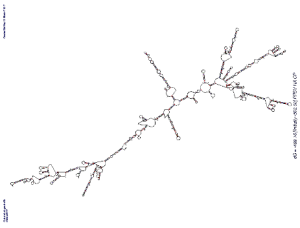
- dsRNA: Most of the described plant viruses have RNA genomes and one of the byproducts of viral replication is double-stranded RNA (dsRNA)5. This dsRNA is very stable and triggers the RNAi pathway. However, even with DNA viruses, some of the genes overlap and can trigger the RNAi pathway too6. With the transgenics that utilized the complete coat protein but triggered the RNAi pathway, it was found that in some cases the secondary structure of the RNA produced regions with dsRNA that could trigger RNA silencing in plants (see image at right)7.
- Short hairpins: The discovery that short dsRNA segments could trigger RNAi led to the use of constructs that generate a hairpin cassette that forms into dsRNA8. Early uses of virus-derived transgenic resistance used entire genes; however, with advances in our understanding of the RNAi pathway, many researchers have adopted the hairpin cassette method as it allows for the targeting of multiple viruses and/or viral genes9. This has several benefits. By targeting multiple genes, researchers are able to minimize the chances of the targeted virus developing resistance to the transgenic plant. The hairpin cassette technique also allows researchers to target multiple viruses with a single construct.
Virus-derived transgenic products on the market
The most widely known transgenic on the market is papaya that is resistant to PRSV10. This product actually saved the papaya industry in Hawaii as PRSV makes fruit unmarketable and eventually kills infected trees. Complicating matters, PRSV is transmitted by several aphids non-persistently (it’s carried in the stylet of the aphid) so it transmits as soon as the aphid probes the plant tissue. Because of this, attempts to control the aphid vectors by spraying does not work as the virus has already been transmitted by the time the aphids are killed.
Resistance to the virus was the only option; however, although resistance to PRSV has been identified in wild relatives, all attempts to introgress the trait were have not been successful introduced into cultivated papaya from a wild relative until just recently11 after 50 years of attempts by plant breeders. The only strategy that papaya growers could use was moving their operations to another island. Each time they moved, there was a short reprieve, but the virus eventually made it to that area. To combat this, work was started to investigate the potential for cross protection; however, it was not effective with the isolates found in Hawaii. Luckily at around that time, news of a new technique was announced that used a transgenic with the coat protein of a virus that provided resistance to that virus. So work on a transgenic began. By the time PRSV made it to the last papaya growing area in Puna, Hawaii, the transgenic was ready and the industry was saved.

There are two different transgenic events for virus resistance in summer squash. The first, ZW-2012, targets ZYMV and WMV whereas the second, CZW-313, targets CMV in addition to ZYMV and WMV. ZYMV and WMV are related to PRSV (they are all in the same genus, Potyvirus) and are targeted by introduction of the entire coat protein gene that induces the RNAi pathway in plants. The way that the CMV construct works is interesting. It seems that the introduction of the coat protein gene acts by interfering with the life cycle of the virus and by inducing the RNAi pathway.
Virus-derived transgenic products in development
There are a number of transgenic crops that utilize virus-derived resistance currently in development. These range from rice plants engineered to resist Rice grassy stunt tenuivirus14 to cotton plants that are resistant to Cotton leaf curl Kokhran begomovirus15 to tomatoes resistant to Tomato yellow leaf curl begomovirus16 to lettuce that is resistant to Mirafiori lettuce big-vein ophiovirus17. (See here for an article on Biofortified about virus-resistant black beans in Brazil.) Each of these viruses causes severe losses in their respective crops and durable resistance either has not been found or has been hard to introduce into the respective crop plants. This is just a small subset of the virus-derived transgenic plants that are in various stages of development worldwide. However, there are several viral diseases that cause food insecurity and/or severe economic losses that have transgenic solutions. Two that are threats to food security will be discussed below:
Cassava mosaic disease (CMD)

This disease contributes to food insecurity in Africa and Southern Asia18 and is caused by at least 10 related virus species in the genus Begomovirus. Losses to CMD are due to the absence of tubers on infected plants. Since cassava grows well in poor soil with less rain than other staple crops, it is widely grown in sub-Saharan Africa. Because of the severity of the losses and because cassava is often a crop counted on to help reduce the effects of famine, it is an ideal target for virus-derived transgenic resistance.
By targeting a gene that is essential for begomovirus replication (AC1), researchers were able to generate cassava plants that were resistant19 to African cassava mosaic begomovirus as well as two related cassava begomoviruses. This was then improved further by targeting two additional genes essential for begomovirus replication (AC2 and AC3)20. The effort to develop virus-derived transgenic resistance to CMD has been included in the VIRCA Plus project which combines resistance to CMD, cassava brown streak disease and nutritional improvements (transgenic fortification with zinc and iron) with to cassava.
Cassava brown streak disease (CBSD)
Like CMD, CBSD is a threat to food security. Although CBSD can reduce tuber size it does not result in no tubers as with CMD. However, what remains of CBSD symptomatic tubers are inedible due to necrosis in the tubers making infected crops a near total loss21. CBSD is caused by viruses that are transmitted by whiteflies. However, there are only two known species, Cassava brown streak ipomovirus and Ugandan cassava brown streak ipomovirus, and both are generally just referred to as CBSV. Like CMD, the viruses that cause CSBD have been targeted using virus-derived transgenic resistance22 and as mentioned above, transgenic resistance to these viruses have been included in the VIRCA Plus project.
Other avenues for using RNA silencing
The usefulness of RNA silencing is not limited to resistance to plant viruses. There have been other examples of this technology being used to prevent browning due to oxidation (Arctic apple and the Simplot Innate potatoes – see articles on Biofortified about the apple and potato) and reducing acrylamide formation in cooked potatoes (also the Innate potato). Another area being explored that relates to transgenic virus resistance is the development of RNA silencing for arthropod vectors of plant viruses. This is still in the early stages of development, but shows great promise in offering more options for farmers to use in integrated pest management. As with virus-derived resistance, RNA silencing for insect control has focused on turning off genes that are essential for the insect to live, such as v-ATPase subunit A in whiteflies23, or genes that are essential for insect-plant interactions, such as C002 which is expressed in the salivary glands of aphids24. There will be an additional post discussing the use of RNA silencing for other uses in the near future.
Conclusions
Virus-derived transgenic resistance holds great promise in sparing growers and consumers the costs of losses due to virus infection. Furthermore, this technology has saved at least one crop, papaya grown in Hawaii, and holds the potential to grant those in developing nations food security by preventing losses in staple crops. Some of the other benefits of this approach to controlling plant viruses is that it reduces sprays that were used to control the arthropod vectors, while not altering how the crops are grown. One of the main challenges is that resistance to one strain of virus may not give strong resistance to other strains, so the evolution of new virus strains must be closely monitored.
References:
- Leitner et al., 2015. Arthropod Vectors and Disease Transmission: Translational Aspects. PLoS Neglected Tropical Pathogens 9(11): e0004107. DOI: 10.1371/journal.pntd.0004107
- Whitfield et al., 2015. Insect vector-mediated transmission of plant viruses. Virology Volumes 479–480: 278–289 DOI: 10.1016/j.virol.2015.03.026
- Beachy, 1999. Coat-protein-mediated resistance to tobacco mosaic virus: discovery mechanisms and exploitation. Philos Trans R Soc Lond B Biol Sci 354:659-664. DOI: 10.1098/rstb.1999.0418
- Obbard et al., 2009. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci 364(1513): 99–115. DOI: 10.1098/rstb.2008.0168
- Weber et al., 2006. Double-Stranded RNA Is Produced by Positive-Strand RNA Viruses and DNA Viruses but Not in Detectable Amounts by Negative-Strand RNA Viruses. Journal of Virology. 80(10): 5059–5064. DOI: 10.1128/JVI.80.10.5059-5064.2006
- Li et al., 2014. Suppression of RNA Silencing by a Plant DNA Virus Satellite Requires a Host Calmodulin-Like Protein to Repress RDR6 Expression. PLoS One. DOI: 10.1371/journal.ppat.1003921
- Lindbo and Falk, 2017. The Impact of “Coat Protein-Mediated Virus Resistance” in Applied Plant Pathology and Basic Research. Phytopathology 107(6): 624-634 DOI: 10.1094/PHYTO-12-16-0442-RVW
- Jia et al., 2007. A strategy for constructing and verifying short hairpin RNA expression vectors. J RNAi Gene Silencing 3(1): 248–253. PMCID: PMC2737214
- Lambeth et al., 2010. A direct comparison of strategies for combinatorial RNA interference. BMC Molecular Biology 11:77. DOI: 10.1186/1471-2199-11-77
- Gonsalves et al., 2004. Transgenic Virus Resistant Papaya: From Hope to Reality for Controlling Papaya Ringspot Virus in Hawaii. APSnet Features. Online. DOI: 10.1094/APSnetFeature-2004-0704
- Siar et al., 2011. Papaya ringspot virus resistance in Carica papaya via introgression from Vasconcellea quercifolia. Euphytica 181: 159-168 DOI: 10.1007/s10681-011-0388-z
- Fuchs and Gonsalves, 1995. Resistance of Transgenic Hybrid Squash ZW-20 Expressing the Coat Protein Genes of Zucchini Yellow Mosaic Virus and Watermelon Mosaic Virus 2 to Mixed Infections by Both Potyviruses. Nature Biotechnology 13: 1466 – 1473 DOI: 10.1038/nbt1295-1466
- Tricoll et al., 1995. Field Evaluation of Transgenic Squash Containing Single or Multiple Virus Coat Protein Gene Constructs for Resistance to Cucumber Mosaic Virus, Watermelon Mosaic Virus 2, and Zucchini Yellow Mosaic Virus. Nature Biotechnology 13: 1458 – 1465 DOI: 10.1038/nbt1295-1458
- Shimizu et al., 2013. Strong Resistance Against Rice grassy stunt virus Is Induced in Transgenic Rice Plants Expressing Double-Stranded RNA of the Viral Genes for Nucleocapsid or Movement Proteins as Targets for RNA Interference. Phytopathology 103: 513-519 DOI: 10.1094/PHYTO-07-12-0165-R
- Yasmeen et al., 2016. Amplicon-Based RNA Interference Targeting V2 Gene of Cotton Leaf Curl Kokhran Virus-Burewala Strain Can Provide Resistance in Transgenic Cotton Plants. Molecular Biotechnology 58: 807-820 DOI: 10.1007/s12033-016-9980-8
- Fuentes et al., 2016. Field Trial and Molecular Characterization of RNAi-Transgenic Tomato Plants That Exhibit Resistance to Tomato Yellow Leaf Curl Geminivirus. Molecular Plant-Microbe Interactions 29: 197-209 DOI: 10.1094/MPMI-08-15-0181-R
- Kawazu et al., 2016. Development of marker-free transgenic lettuce resistant to Mirafiori lettuce big-vein virus. Transgenic Research 25: 711-719 DOI: 10.1007/s11248-016-9956-2
- Alabi et al., 2011. Cassava Mosaic Disease: A Curse to Food Security in Sub-Saharan Africa. APSnet Features. Online. DOI: 10.1094/APSnetFeature-2011-0701
- Chellappan et al., 2004. Broad Spectrum Resistance to ssDNA Viruses Associated with Transgene-Induced Gene Silencing in Cassava. Plant Molecular Biology 56: 601-611 DOI: 10.1007/s11103-004-0147-9
- Zhang et al., 2005. Resistance to cassava mosaic disease in transgenic cassava expressing antisense RNAs targeting virus replication genes. Plant Biotechnology Journal 3: 385-397 DOI: 10.1111/j.1467-7652.2005.00132.x
- Patil et al., 2015. Cassava brown streak disease: a threat to food security in Africa. Journal of General Virology 96: 956-968 DOI: 10.1099/vir.0.000014
- Ogwok et al., 2012. Transgenic RNA interference (RNAi)-derived field resistance to cassava brown streak disease. Molecular Plant Pathology 13: 1019-1031 DOI: 10.1111/j.1364-3703.2012.00812.x
- Thakur et al., 2014. Enhanced Whitefly Resistance in Transgenic Tobacco Plants Expressing Double Stranded RNA of v-ATPase A Gene. PLoS One 9(3): e87235 DOI: 10.1371/journal.pone.0087235
- Pitino et al., 2011. Silencing of Aphid Genes by dsRNA Feeding from Plants. PLoS One 6(10): e25709 DOI: 10.1371/journal.pone.0025709
Written by Guest Expert
Alma Laney works with plant viruses and the arthropods who vector them and on his blog The Mad Virologist he covers all aspects of virology from human pathogens to archaea viruses and everything in between. http://themadvirologist.blogspot.com/

